Tidy Tuesday - Coffee Rating
Predicting Process of Green Coffee Beans
With coffee being a hobby of mine, I was scrolling through past Tidy Tuesdays and found one on coffee ratings. Originally I thought looking at predictions of total cup points, but I assumed with all the coffee tasting characteristics that it wouldn’t really tell me anything. Instead, I decided to look into the processing method, as there are different taste characteristics between washed and other processing methods.
coffee <- read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2020/2020-07-07/coffee_ratings.csv') %>%
mutate(species = as.factor(species),
process = recode(processing_method, "Washed / Wet" = "washed",
"Semi-washed / Semi-pulped" = "not_washed",
"Pulped natural / honey" = "not_washed",
"Other" = "not_washed",
"Natural / Dry" = "not_washed",
"NA" = NA_character_),
process = as.factor(process),
process = relevel(process, ref = "washed"),
country_of_origin = as.factor(country_of_origin)) %>%
drop_na(process) %>%
filter(country_of_origin != "Cote d?Ivoire")
##
## -- Column specification --------------------------------------------------------
## cols(
## .default = col_character(),
## total_cup_points = col_double(),
## number_of_bags = col_double(),
## aroma = col_double(),
## flavor = col_double(),
## aftertaste = col_double(),
## acidity = col_double(),
## body = col_double(),
## balance = col_double(),
## uniformity = col_double(),
## clean_cup = col_double(),
## sweetness = col_double(),
## cupper_points = col_double(),
## moisture = col_double(),
## category_one_defects = col_double(),
## quakers = col_double(),
## category_two_defects = col_double(),
## altitude_low_meters = col_double(),
## altitude_high_meters = col_double(),
## altitude_mean_meters = col_double()
## )
## i Use `spec()` for the full column specifications.
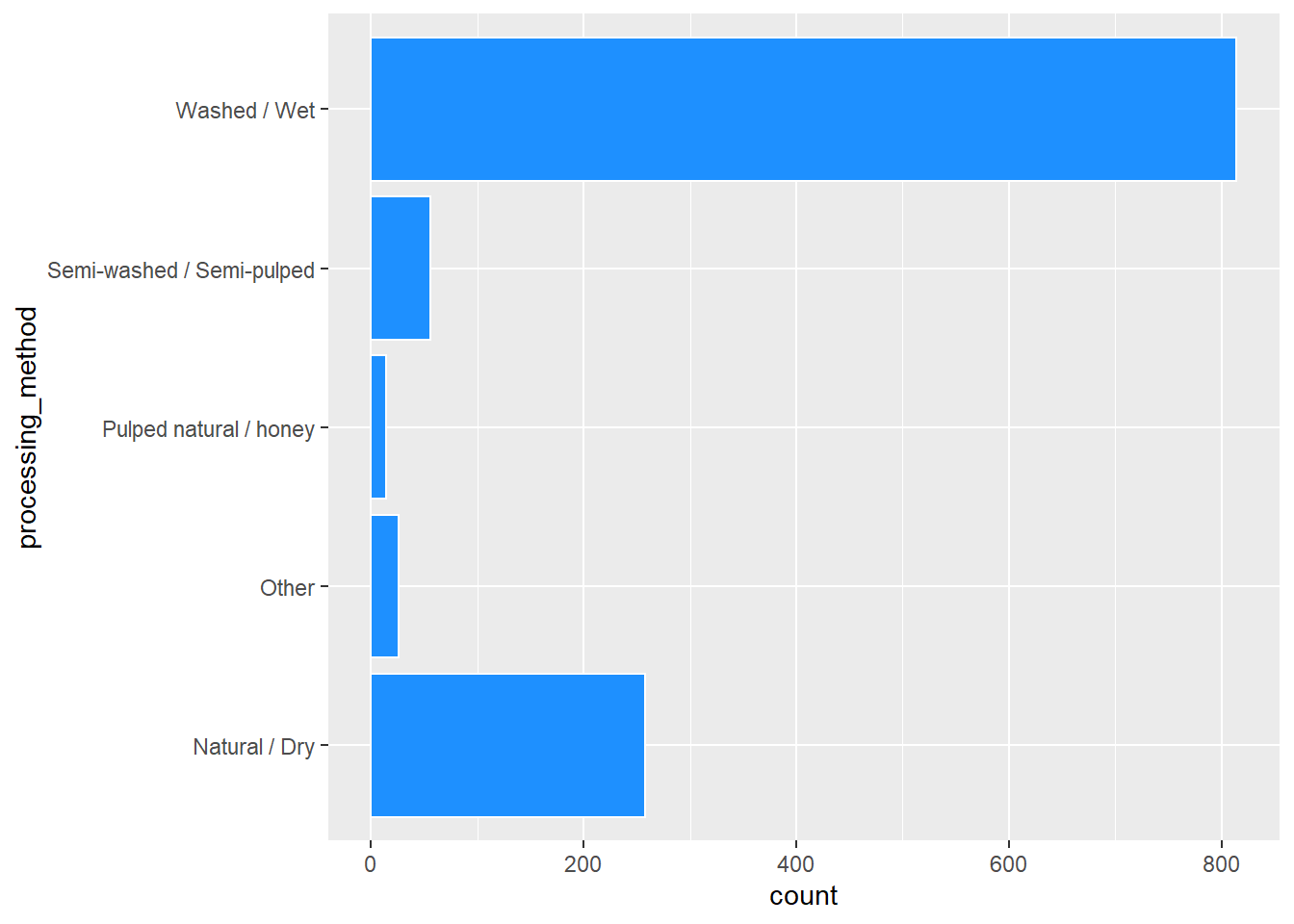
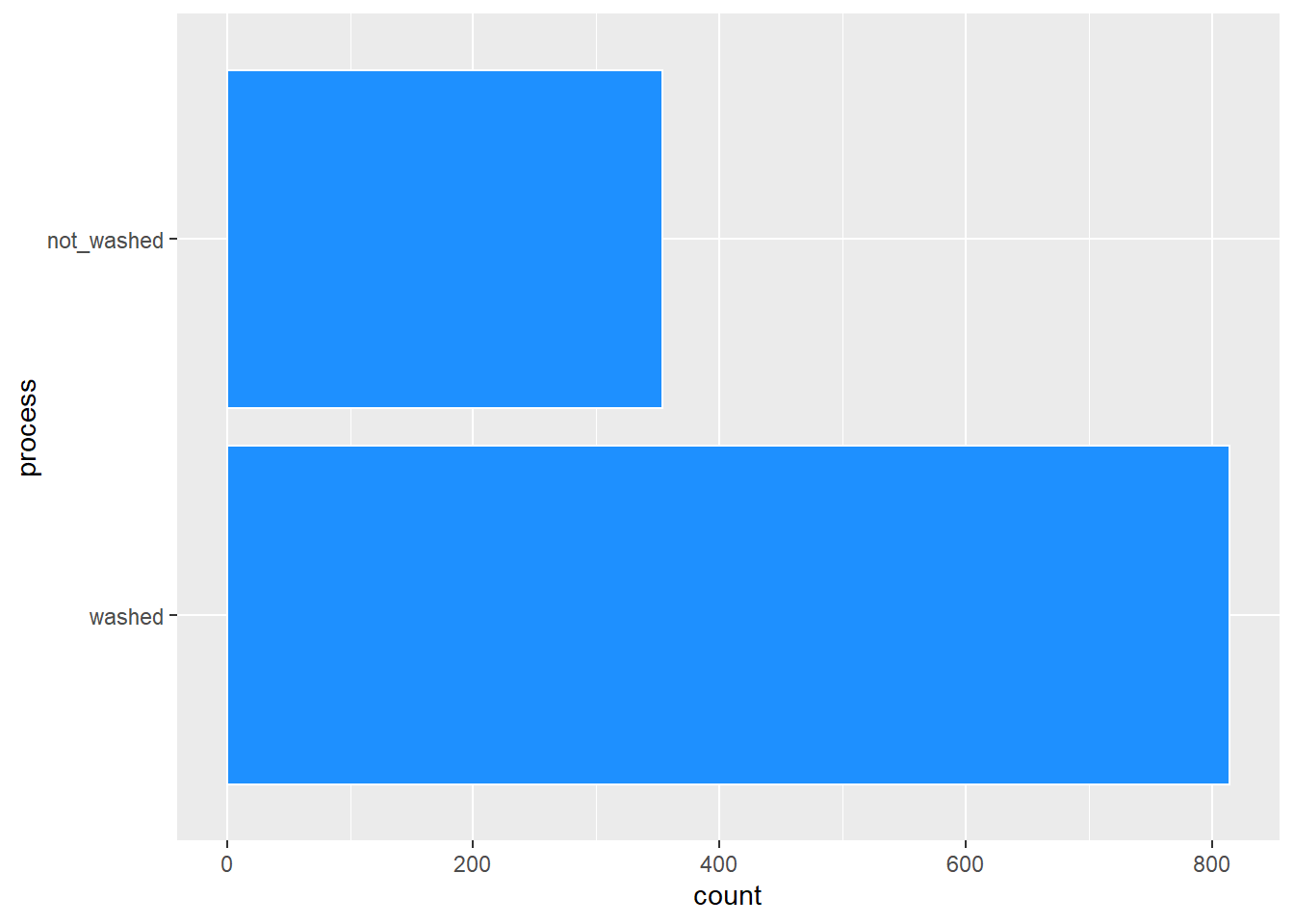
After looking at the distributions of procssing methods, I also decided to make the processing method binary with washed and not washed. This worked out better for the prediction models. There are also some descriptives of each variable.
coffee %>%
ggplot(aes(processing_method)) +
geom_bar(color = "white", fill = "dodgerblue") +
coord_flip()
coffee %>%
ggplot(aes(process)) +
geom_bar(color = "white", fill = "dodgerblue") +
coord_flip()
psych::describe(coffee, na.rm = TRUE)[c("n", "mean", "sd", "min", "max", "skew", "kurtosis")]
## n mean sd min max skew kurtosis
## total_cup_points 1168 82.06 2.71 59.83 90.58 -1.95 9.26
## species* 1168 1.01 0.09 1.00 2.00 10.65 111.61
## owner* 1161 133.43 76.78 1.00 287.00 0.10 -1.00
## country_of_origin* 1168 14.79 10.08 1.00 36.00 0.31 -1.12
## farm_name* 883 271.04 154.16 1.00 524.00 0.00 -1.27
## lot_number* 239 93.18 59.86 1.00 202.00 0.18 -1.27
## mill* 931 196.60 122.71 1.00 419.00 0.24 -1.28
## ico_number* 1057 360.89 242.05 1.00 753.00 0.08 -1.27
## company* 1077 141.73 76.77 1.00 266.00 -0.10 -1.23
## altitude* 1014 154.54 98.40 1.00 351.00 0.32 -1.04
## region* 1137 174.91 87.74 1.00 325.00 -0.11 -1.09
## producer* 995 314.12 175.27 1.00 624.00 -0.04 -1.12
## number_of_bags 1168 153.80 130.08 1.00 1062.00 0.37 0.50
## bag_weight* 1168 23.50 16.67 1.00 45.00 -0.23 -1.71
## in_country_partner* 1168 9.68 7.04 1.00 25.00 0.41 -1.31
## harvest_year* 1161 5.82 2.95 1.00 14.00 0.76 -0.46
## grading_date* 1168 242.39 144.52 1.00 495.00 0.11 -1.21
## owner_1* 1161 135.29 77.68 1.00 290.00 0.09 -1.00
## variety* 1089 12.61 9.77 1.00 29.00 0.63 -1.24
## processing_method* 1168 3.98 1.67 1.00 5.00 -1.13 -0.62
## aroma 1168 7.56 0.31 5.08 8.75 -0.55 4.46
## flavor 1168 7.51 0.34 6.08 8.83 -0.34 1.73
## aftertaste 1168 7.39 0.34 6.17 8.67 -0.45 1.36
## acidity 1168 7.53 0.31 5.25 8.75 -0.30 3.31
## body 1168 7.52 0.28 6.33 8.50 -0.10 0.89
## balance 1168 7.51 0.34 6.08 8.58 -0.10 1.17
## uniformity 1168 9.84 0.50 6.00 10.00 -4.21 20.83
## clean_cup 1168 9.84 0.75 0.00 10.00 -6.98 62.29
## sweetness 1168 9.89 0.52 1.33 10.00 -7.53 80.78
## cupper_points 1168 7.48 0.40 5.17 8.75 -0.64 2.79
## moisture 1168 0.09 0.05 0.00 0.17 -1.41 0.35
## category_one_defects 1168 0.51 2.70 0.00 63.00 14.43 279.42
## quakers 1167 0.17 0.82 0.00 11.00 6.87 57.30
## color* 1070 2.80 0.64 1.00 4.00 -1.51 2.57
## category_two_defects 1168 3.79 5.54 0.00 55.00 3.54 18.53
## expiration* 1168 241.61 144.11 1.00 494.00 0.12 -1.21
## certification_body* 1168 9.38 6.65 1.00 24.00 0.37 -1.31
## certification_address* 1168 16.58 7.33 1.00 29.00 -0.19 -1.06
## certification_contact* 1168 9.39 7.26 1.00 26.00 0.36 -0.96
## unit_of_measurement* 1168 1.87 0.34 1.00 2.00 -2.21 2.87
## altitude_low_meters 1012 1796.86 9073.21 1.00 190164.00 19.36 384.12
## altitude_high_meters 1012 1834.27 9071.86 1.00 190164.00 19.36 384.03
## altitude_mean_meters 1012 1815.56 9072.31 1.00 190164.00 19.36 384.12
## process* 1168 1.30 0.46 1.00 2.00 0.86 -1.27
Now, its time to split the data into training and testing data. I also included the function of strata to stratify sampling based on process.
set.seed(05132021)
coffee_split <- initial_split(coffee, strata = "process")
coffee_train <- training(coffee_split)
coffee_test <- testing(coffee_split)
I also did some cross validation for the training dataset and used the metrics I was most interested in.
set.seed(05132021)
coffee_fold <- vfold_cv(coffee_train, strata = "process", v = 10)
metric_measure <- metric_set(accuracy, mn_log_loss, roc_auc)
From the beginning I was interested in the tasting characteristics and how they would predict whether the green coffee was washed or not washed. I also included the total cup points because I wanted to see the importance of that predictor on the processing method. The only feature engineering I did was to remove any zero variance in the predictors of the model.
set.seed(05132021)
char_recipe <- recipe(process ~ aroma + flavor + aftertaste +
acidity + body + balance + uniformity + clean_cup +
sweetness + total_cup_points,
data = coffee_train) %>%
step_zv(all_predictors(), -all_outcomes())
char_recipe %>%
prep() %>%
bake(new_data = NULL) %>%
head()
## # A tibble: 6 x 11
## aroma flavor aftertaste acidity body balance uniformity clean_cup sweetness
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 8.17 8.58 8.42 8.42 8.5 8.25 10 10 10
## 2 8.08 8.58 8.5 8.5 7.67 8.42 10 10 10
## 3 8.17 8.17 8 8.17 8.08 8.33 10 10 10
## 4 8.42 8.17 7.92 8.17 8.33 8 10 10 10
## 5 8.5 8.5 8 8 8 8 10 10 10
## 6 8 8 8 8.25 8 8.17 10 10 10
## # ... with 2 more variables: total_cup_points <dbl>, process <fct>
Logistic Regression
The first model I wanted to test with the current recipe was logistic regression. The accuracy and roc auc were alright for a starting model.
## Warning: package 'rlang' was built under R version 4.0.4
collect_metrics(lr_fit)
## # A tibble: 3 x 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.698 10 0.00342 Preprocessor1_Model1
## 2 mn_log_loss binary 0.589 10 0.00775 Preprocessor1_Model1
## 3 roc_auc binary 0.648 10 0.0188 Preprocessor1_Model1
Lasso Regression
Now for the first penalized regression. The lasso regression did not improve in either metric. Let’s try the next penalized regression.
collect_metrics(lasso_fit)
## # A tibble: 3 x 6
## .metric .estimator mean n std_err .config
## <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 accuracy binary 0.702 10 0.00397 Preprocessor1_Model1
## 2 mn_log_loss binary 0.592 10 0.00850 Preprocessor1_Model1
## 3 roc_auc binary 0.645 10 0.0191 Preprocessor1_Model1
Ridge Regression
The ridge regression was shown to not be a good fitting model. So I tested an additional penalized regression while tuning hyper-parameters.
collect_metrics(ridge_fit)
Elastic Net Regression
The elastic net regression had slightly better accuracy than the non-penalized logistic regression but the ROC AUC was exactly the same. While the elastic net regression did not take long computationally due to the small amount of data, this model would not be chosen over the logistic regression.
collect_metrics(elastic_fit)
## # A tibble: 300 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 1 e-10 0 accuracy binary 0.698 10 0.00342 Preprocessor1_~
## 2 1 e-10 0 mn_log_lo~ binary 0.589 10 0.00775 Preprocessor1_~
## 3 1 e-10 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_~
## 4 1.29e- 9 0 accuracy binary 0.698 10 0.00342 Preprocessor1_~
## 5 1.29e- 9 0 mn_log_lo~ binary 0.589 10 0.00775 Preprocessor1_~
## 6 1.29e- 9 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_~
## 7 1.67e- 8 0 accuracy binary 0.698 10 0.00342 Preprocessor1_~
## 8 1.67e- 8 0 mn_log_lo~ binary 0.589 10 0.00775 Preprocessor1_~
## 9 1.67e- 8 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_~
## 10 2.15e- 7 0 accuracy binary 0.698 10 0.00342 Preprocessor1_~
## # ... with 290 more rows
show_best(elastic_fit, metric = "accuracy", n = 5)
## # A tibble: 5 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 7.74e- 2 0.111 accuracy binary 0.703 10 0.00434 Preprocessor1_Mo~
## 2 1 e-10 0.333 accuracy binary 0.703 10 0.00422 Preprocessor1_Mo~
## 3 1.29e- 9 0.333 accuracy binary 0.703 10 0.00422 Preprocessor1_Mo~
## 4 1.67e- 8 0.333 accuracy binary 0.703 10 0.00422 Preprocessor1_Mo~
## 5 2.15e- 7 0.333 accuracy binary 0.703 10 0.00422 Preprocessor1_Mo~
show_best(elastic_fit, metric = "roc_auc", n = 5)
## # A tibble: 5 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 1 e-10 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_Mod~
## 2 1.29e- 9 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_Mod~
## 3 1.67e- 8 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_Mod~
## 4 2.15e- 7 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_Mod~
## 5 2.78e- 6 0 roc_auc binary 0.648 10 0.0188 Preprocessor1_Mod~
select_best(elastic_fit, metric = "accuracy")
## # A tibble: 1 x 3
## penalty mixture .config
## <dbl> <dbl> <chr>
## 1 0.0774 0.111 Preprocessor1_Model019
select_best(elastic_fit, metric = "roc_auc")
## # A tibble: 1 x 3
## penalty mixture .config
## <dbl> <dbl> <chr>
## 1 0.0000000001 0 Preprocessor1_Model001
New Recipe
Even though the elastic net regression was only slightly better, I decided to update the workflow using that model. This time I decided to update the recipe by including additional predictors like if there were any defects in the green coffee beans, the species of the coffee (e.g., Robusta and Arabica), and the country of origin. I also included additional steps in my recipe by transforming the category predictors and working with the factor predictors, like species, and country of origin. The inclusion of additional steps and the predictors created a better fitting model with the elastic net regression.
set.seed(05132021)
bal_rec <- recipe(process ~ aroma + flavor + aftertaste +
acidity + body + balance + uniformity + clean_cup +
sweetness + total_cup_points + category_one_defects + category_two_defects + species +
country_of_origin,
data = coffee_train) %>%
step_BoxCox(category_two_defects, category_one_defects) %>%
step_novel(species, country_of_origin) %>%
step_other(species, country_of_origin, threshold = .01) %>%
step_unknown(species, country_of_origin) %>%
step_dummy(species, country_of_origin) %>%
step_zv(all_predictors(), -all_outcomes())
collect_metrics(elastic_bal_fit)
## # A tibble: 300 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 1 e-10 0 accuracy binary 0.839 10 0.00923 Preprocessor1_~
## 2 1 e-10 0 mn_log_lo~ binary 0.431 10 0.0153 Preprocessor1_~
## 3 1 e-10 0 roc_auc binary 0.840 10 0.0163 Preprocessor1_~
## 4 1.29e- 9 0 accuracy binary 0.839 10 0.00923 Preprocessor1_~
## 5 1.29e- 9 0 mn_log_lo~ binary 0.431 10 0.0153 Preprocessor1_~
## 6 1.29e- 9 0 roc_auc binary 0.840 10 0.0163 Preprocessor1_~
## 7 1.67e- 8 0 accuracy binary 0.839 10 0.00923 Preprocessor1_~
## 8 1.67e- 8 0 mn_log_lo~ binary 0.431 10 0.0153 Preprocessor1_~
## 9 1.67e- 8 0 roc_auc binary 0.840 10 0.0163 Preprocessor1_~
## 10 2.15e- 7 0 accuracy binary 0.839 10 0.00923 Preprocessor1_~
## # ... with 290 more rows
show_best(elastic_bal_fit, metric = "accuracy", n = 5)
## # A tibble: 5 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.000464 0.111 accuracy binary 0.840 10 0.0101 Preprocessor1_Model0~
## 2 0.000464 0.222 accuracy binary 0.840 10 0.0101 Preprocessor1_Model0~
## 3 0.00599 0.111 accuracy binary 0.840 10 0.00933 Preprocessor1_Model0~
## 4 0.00599 0.222 accuracy binary 0.840 10 0.00933 Preprocessor1_Model0~
## 5 0.00599 0.333 accuracy binary 0.840 10 0.00933 Preprocessor1_Model0~
show_best(elastic_bal_fit, metric = "mn_log_loss", n = 5)
## # A tibble: 5 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.000464 1 mn_log_lo~ binary 0.420 10 0.0179 Preprocessor1_Mode~
## 2 0.000464 0.889 mn_log_lo~ binary 0.420 10 0.0178 Preprocessor1_Mode~
## 3 0.000464 0.778 mn_log_lo~ binary 0.420 10 0.0178 Preprocessor1_Mode~
## 4 0.000464 0.667 mn_log_lo~ binary 0.420 10 0.0178 Preprocessor1_Mode~
## 5 0.000464 0.556 mn_log_lo~ binary 0.420 10 0.0177 Preprocessor1_Mode~
show_best(elastic_bal_fit, metric = "roc_auc", n = 5)
## # A tibble: 5 x 8
## penalty mixture .metric .estimator mean n std_err .config
## <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.00599 0.778 roc_auc binary 0.843 10 0.0150 Preprocessor1_Model078
## 2 0.000464 0.667 roc_auc binary 0.843 10 0.0141 Preprocessor1_Model067
## 3 0.00599 0.889 roc_auc binary 0.843 10 0.0148 Preprocessor1_Model088
## 4 0.000464 0.444 roc_auc binary 0.843 10 0.0141 Preprocessor1_Model047
## 5 0.000464 0.556 roc_auc binary 0.843 10 0.0141 Preprocessor1_Model057
select_best(elastic_bal_fit, metric = "accuracy")
## # A tibble: 1 x 3
## penalty mixture .config
## <dbl> <dbl> <chr>
## 1 0.000464 0.111 Preprocessor1_Model017
select_best(elastic_bal_fit, metric = "mn_log_loss")
## # A tibble: 1 x 3
## penalty mixture .config
## <dbl> <dbl> <chr>
## 1 0.000464 1 Preprocessor1_Model097
select_best(elastic_bal_fit, metric = "roc_auc")
## # A tibble: 1 x 3
## penalty mixture .config
## <dbl> <dbl> <chr>
## 1 0.00599 0.778 Preprocessor1_Model078
Now using the testing dataset, we can see how well the final model fit the testing data. While not the best at predicting washed green coffee beans, this was a good test to show that the penalized regressions are not always the best fitting models compared to regular logistic regression. In the end, it seemed like the recipe was the most important component to predicting washed green coffee beans.
final_results %>%
collect_metrics()
## # A tibble: 2 x 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 accuracy binary 0.823 Preprocessor1_Model1
## 2 roc_auc binary 0.817 Preprocessor1_Model1